Abstract
Introduction: Myeloid neoplasia, including acute myeloid leukemia and myelodysplastic syndrome are heterogeneous hematopoietic stem cell disorders which are marked by the acquisition of somatic alterations and clonal evolution 1. Patients with myeloid neoplasia are classified due to the WHO classification systems and besides clinical and hematological criteria, cytogenetic and molecular genetic alterations highly impact treatment stratification 2. In routine diagnostics, a combination of methods is used to decipher different types of genetic variants, i.e. single nucleotide variants (SNVs), insertions/deletions (indels), structural variants (SVs) and copy number variations (CNVs) which may not be detected using one single method.
Methods: We used a bioinformatic approach to analyze clonal evolution and genetic architecture in patients with myeloid neoplasia using single nucleotide variants (SNVs), insertions/deletions (indels), structural variants (SVs) and copy number variations (CNVs). Six patients were comprehensively analyzed using karyotyping, fluorescence in situ hybridization (FISH), array-CGH and a custom NGS panel with 148 genes/ gene regions that are recurrently affected in patients with hematologic neoplasia. At the initial time point or during disease course all patients showed many genomic variants: Two patients (#1, #2) were analyzed at one time point (initial), two patients (#3, #4) were analyzed at two time points (initial and progression), one patient (#5) was analyzed at four time points (initial, progression, remission, relapse), and one patient (#6) was analyzed at five time points (initial, remission, relapse, progression, remission).
Results and Conclusions: Clonal evolution was reconstructed manually, integrating all mutational information on SNVs, indels, SVs and CNVs 3. Cancer cell fractions (CCFs) for SNVs and indels were estimated based on VAFs, assuming heterozygous variants (2*VAF=CCF). CCFs for SVs and CNVs were estimated based on cell counts reported for karyotyping and FISH analyses. For SVs as well as CNVs, which were only detected by array-CGH, CCF was estimated based on logRatio. In case of a CNV overlapping the position of an SNV or indel, calculation of CCF is less straightforward. Altogether, we differentiate between three cases: 1) The CNV occurred prior to the SNV/indel, but in the same cells. 2) The SNV/indel occurred prior to the CNV, but in the same cells. 3) SNV/indel and CNV exist in parallel, independent of each other.
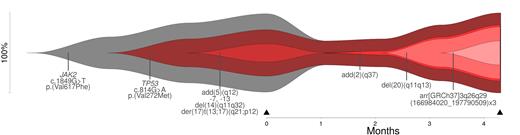
The bioinformatic approach reconstructed clonal evolution (linear and/or branching) for all patients and the results were visualized by fishplots. We identified alterations, which play a role in the pathogenesis of the disease (driver) and alterations, which occur during disease development (passenger). On two samples, we showed that reconstruction of clonal evolution is possible even with data from one time point only. For other samples, providing data on more than one time point, the effect of therapy was estimated.
This bioinformatic approach offers the possibility of analyzing clonal evolution and genetic architecture at one or more time points of analysis. The visualization of the results in fishplots contributes to a better understanding of genetic architecture and helps to identify possible targets for the disease (personalized therapy). Furthermore, this model can be used to identify markers in order to assess minimal residual disease (MRD).
Figure 1 Reconstruction of clonal evolution (time point of analysis: black triangle) for patient #4 (diagnosis: secondary acute myeloid leukemia).
References:
1. Doulatov S, Papapetrou EP. Studying clonal evolution of myeloid malignancies using induced pluripotent stem cells. Curr Opin Hematol. 2021;28(1):50-56.
2. Edited by Swerdlow SH CE, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. International Agency for Research on Cancer. 2017;Revised 4th edition.
3. Reutter K, Sandmann S, Rohde J, et al. Reconstructing clonal evolution in relapsed and non-relapsed Burkitt lymphoma. Leukemia. 2021;35(2):639-643.
Thol: Jazz: Honoraria; BMS/Celgene: Honoraria, Research Funding; Abbvie: Honoraria; Astellas: Honoraria; Novartis: Honoraria; Pfizer: Honoraria. Heuser: Daiichi Sankyo: Membership on an entity's Board of Directors or advisory committees, Research Funding; Bayer Pharma AG: Research Funding; Roche: Membership on an entity's Board of Directors or advisory committees, Research Funding; Novartis: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; BMS/Celgene: Membership on an entity's Board of Directors or advisory committees, Research Funding; Karyopharm: Research Funding; Tolremo: Membership on an entity's Board of Directors or advisory committees; Pfizer: Membership on an entity's Board of Directors or advisory committees, Research Funding; Janssen: Honoraria; Jazz: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Astellas: Research Funding; AbbVie: Membership on an entity's Board of Directors or advisory committees, Research Funding; BergenBio: Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal